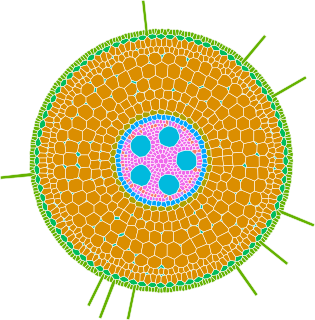
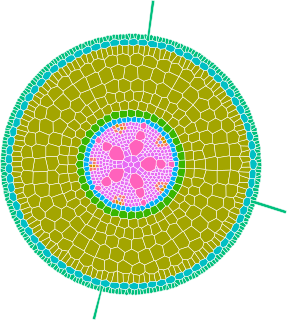
GRANAR is a generator of complete root cross-section network in R.
Upon a small set of anatomical features the GRANAR model is able to reconstruct a generic root cell network for mono and dicotyledon.
Among the few parameters you can modify, there is the aerenchyma proportion, the number and size of the xylem vessels, but also the number of cell layers and the mean size of each cell type.
Once the anatomy is generated, it is possible to look after some functional features related to that anatomy. For instance, hydraulics conductivities can be computed when GRANAR is coupled with MECHA.
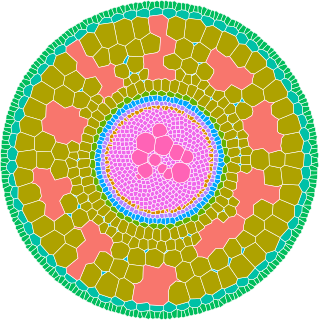
GRANAR is a R package
The source code of GRANAR is available on github: https://github.com/granar/granar
Another github repository contains example parameter files needed to run GRANAR: granar_examples You do not need your own data to try it out.
The first step before running GRANAR is to install a working version for R! Installation of R are available here: https://cran.r-project.org/
GRANAR is built upon a handfull of external libraries. You will need to install them before using the model.
These libraries can be install using RStudio. In the Terminal or Console, enter:
install.packages(deldir) install.packages(alphahull) install.packages(retistruct)
In the Terminal or Console enter the following lines:
install.packages("devtools")
library(devtools)
install_github("granar/granar")
library(granar)
GRANAR uses only parameters files as input. Examples of the input files are stored in the modelparameterfolder in the granar_example directory:
Zea_mays_1_Heymans_2019.xml : General parameters for maize nodal root anatomyGRANAR output are stored in a list element and it can be exported under .xml with the same format as CellSet.
# R script example
library(granar)
params = read_param_xml(path = "params.xml") # import input parameter for GRANAR
sim = list() # create an empty list
sim = create_anatomy(parameters = params) # generate a cell network anatomy
write_anatomy_xml(sim = sim,
path = "new_root.xml") # export the data under a .xml file
GRANAR was developed at the Université catholique de Louvain, in the Earth and Life Insititute, in the lab of Guillaume Lobet. The primary developpers of the model are Adrien Heymans, Marco D'Agostino and Guillaume Lobet. Valentin Couvreur provided some technical assistance.
GRANAR is available at:
GRANAR, a computational tool to better understand the functional importance of monocotyledon root anatomy Adrien Heymans, Valentin Couvreur, Therese LaRue, Ana Paez-Garcia, Guillaume Lobet
Plant Physiology, 2019

